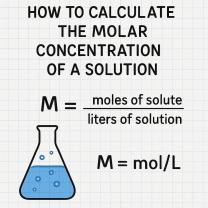
How do I calculate the average CT for my endogenous control?
The average Ct (cycle threshold) for an endogenous control in quantitative real-time polymerase chain reaction (qPCR) is calculated using the Ct values obtained from multiple replicates. The endogenous control, also known as a reference gene, is a gene that should exhibit stable expression across different experimental conditions. Here's a step-by-step process for calculating the average Ct for your endogenous control:
Step 1: Ct Values
- Obtain Ct values: After performing your qPCR experiment, you will have Ct values for each replicate of your endogenous control. The Ct value is the cycle number at which the fluorescence signal crosses a predefined threshold.
Step 2: Identify Replicates
- Identify replicates: Ensure you have multiple replicates (technical or biological) for your endogenous control. The more replicates you have, the more robust your average calculation will be.
Step 3: Calculate Average Ct
Add Ct values: Sum the Ct values for all replicates of your endogenous control.
Divide by the number of replicates: Divide the sum by the total number of replicates.
Example:
Suppose you have three replicates with Ct values of 20.5, 21, and 19.5 for your endogenous control.
So, the average Ct for your endogenous control is 20.33.
Note:
- A lower Ct value indicates higher expression, and a higher Ct value indicates lower expression.
- The stability of the endogenous control's expression across different samples is crucial for accurate normalization of gene expression data.
Always consider best practices in qPCR experimental design, including proper controls, validation of reference genes, and appropriate replicates, to ensure the reliability of your results. If possible, consult with a molecular biologist or a bioinformatician for guidance on your specific experimental setup.
Sure, I can help you with that.
How do you calculate the average CT for an endogenous control?
The average CT (cycle threshold) for an endogenous control is calculated by taking the mean of the CT values for that gene across all samples. This value is then used to normalize the expression of other genes in the experiment.
Here are the steps on how to calculate the average CT for an endogenous control:
- Collect the CT values for the endogenous control gene from all samples.
- Calculate the mean of the CT values.
- The mean CT value is the average CT for the endogenous control gene.
What significance does the average CT hold in scientific experiments?
The average CT for an endogenous control is important because it helps to control for variations in the amount of input RNA between samples. This can be due to factors such as differences in sample quality, RNA extraction efficiency, and cDNA synthesis efficiency.
By normalizing gene expression data to the average CT of an endogenous control, researchers can compare gene expression levels across different samples more accurately. This is important for a variety of scientific experiments, such as those that involve comparing gene expression levels between different cell types, tissues, or conditions.
What methods are used to determine the average CT for an endogenous control?
There are a few different methods that can be used to determine the average CT for an endogenous control. The most common method is to simply calculate the mean of the CT values for all samples. However, there are also more sophisticated methods that can be used, such as:
- The ΔΔCT method: This method involves calculating the difference in CT values between the endogenous control gene and the gene of interest. This difference is then normalized to the difference in CT values between the endogenous control gene and a reference gene.
- The Pfaffl method: This method is similar to the ΔΔCT method, but it uses a different formula to calculate the difference in CT values.
The best method to use for determining the average CT for an endogenous control will depend on the specific experiment and the type of data being analyzed.
I hope this helps! Is there anything else I can help you with?